The Molecular Genetics and Integrative Physiology of Host Defense in C. elegans
Interactions between microbes and host organisms—symbiotic, pathogenic, or commensal—have shaped the evolution of multicellular life. We are interested in the mechanisms of how microbes influence host physiology. We investigate these processes in Caenorhabditis elegans, a simple animal host. We take a molecular genetic approach to the study of host physiology, including behavioral responses of C. elegans to bacteria, and the evolutionarily conserved signaling nexus of innate immunity, stress physiology, and aging.
Research Summary
We are broadly interested in understanding how the physiology 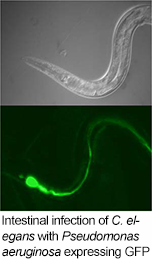 and evolution of multicellular organisms is shaped by their interaction with microbes. Our experimental system is focused on Caenorhabditis elegans, which we were motivated to use in our studies because of the powerful molecular genetic tools that have been developed to study biology in this simple animal host organism. We started with a forward genetic approach to identify genes required for host survival during microbial infection, identifying a role for evolutionarily conserved innate immune signaling pathways in C. elegans defense against pathogenic bacteria (Kim et al., 2002). The genetic analysis of host defense of C. elegans, using both standard laboratory strains as well as strains of C. elegans isolated from the wild, has led to the identification and characterization of multiple modes of host defense, including innate immunity (Troemel et al., 2006; Shivers et al., 2010; Youngman et al., 2011), host tolerance to the stress induced by infection (Richardson et al., 2010; Richardson et al., 2011), and behavioral avoidance (Reddy et al., 2009; Shivers et al., 2009; Reddy et al., 2011; Chang et al., 2011).
and evolution of multicellular organisms is shaped by their interaction with microbes. Our experimental system is focused on Caenorhabditis elegans, which we were motivated to use in our studies because of the powerful molecular genetic tools that have been developed to study biology in this simple animal host organism. We started with a forward genetic approach to identify genes required for host survival during microbial infection, identifying a role for evolutionarily conserved innate immune signaling pathways in C. elegans defense against pathogenic bacteria (Kim et al., 2002). The genetic analysis of host defense of C. elegans, using both standard laboratory strains as well as strains of C. elegans isolated from the wild, has led to the identification and characterization of multiple modes of host defense, including innate immunity (Troemel et al., 2006; Shivers et al., 2010; Youngman et al., 2011), host tolerance to the stress induced by infection (Richardson et al., 2010; Richardson et al., 2011), and behavioral avoidance (Reddy et al., 2009; Shivers et al., 2009; Reddy et al., 2011; Chang et al., 2011).
Our studies in these areas have broadened our perspectives and developed into three principal areas of current interest: 1) Genetic analysis of chemosensory signaling mechanisms and neuroendocrine signals involved in microbial recognition (Meisel et al., 2014)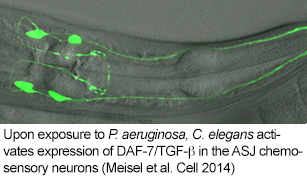 , 2) Cell-autonomous and cell-non-autonomous mechanisms of stress signaling and homeostasis in development, immunity, and aging (Kulalert et al., 2013), and 3) The identification and characterization of small molecules, derived from both host and microbe, that mediate communication between host and microbe and may serve as the basis for the development of novel antimicrobial agents (in collaboration with F. Schroeder at Cornell U.).
, 2) Cell-autonomous and cell-non-autonomous mechanisms of stress signaling and homeostasis in development, immunity, and aging (Kulalert et al., 2013), and 3) The identification and characterization of small molecules, derived from both host and microbe, that mediate communication between host and microbe and may serve as the basis for the development of novel antimicrobial agents (in collaboration with F. Schroeder at Cornell U.).
 and evolution of multicellular organisms is shaped by their interaction with microbes. Our experimental system is focused on Caenorhabditis elegans, which we were motivated to use in our studies because of the powerful molecular genetic tools that have been developed to study biology in this simple animal host organism. We started with a forward genetic approach to identify genes required for host survival during microbial infection, identifying a role for evolutionarily conserved innate immune signaling pathways in C. elegans defense against pathogenic bacteria (
and evolution of multicellular organisms is shaped by their interaction with microbes. Our experimental system is focused on Caenorhabditis elegans, which we were motivated to use in our studies because of the powerful molecular genetic tools that have been developed to study biology in this simple animal host organism. We started with a forward genetic approach to identify genes required for host survival during microbial infection, identifying a role for evolutionarily conserved innate immune signaling pathways in C. elegans defense against pathogenic bacteria ( , 2) Cell-autonomous and cell-non-autonomous mechanisms of stress signaling and homeostasis in development, immunity, and aging (
, 2) Cell-autonomous and cell-non-autonomous mechanisms of stress signaling and homeostasis in development, immunity, and aging (